Abstract
We wrote about generative neural networks in two previous blog posts where we promised to return to the topic in a future update. This is it. This article is a review of some of more advances in autoencoders over the last 10 years. We present examples of denoising autoencoders, variational and three different adversarial neural networks. The presentation is not theoretical, and it uses examples via Jupyter notebooks that can be run on a standard laptop.
Introduction
Autoencoders are a class of deep neural networks that can learn efficient representations of large data collections. The representation is required to be robust enough to regenerate a good approximation of the original data. This can be useful for removing nose from the data when the noise is not an intrinsic property of the underlying data. These autoencoder often, but not always, work by projecting the data into a lower dimensional space (much like what a principal component analysis achieves). We will look at an example of a “denoising autoencoder” below where the underlying signal is identified through a series of convolutional filters.
A difference class of autoencoders are called Generative Autoencoders that have the property that they can create a mathematical machine that can reproduce the probability distribution of the essential features of a data collection. In other words, a trained generative autoencoder can create new instances of ‘fake’ data that has the same statistical properties as the training data. Having a new data source, albeit and artificial one, can be very useful in many studies where it is difficult to find additional examples that fit a given profile. This ideas is being used in research in particle physics to improve the search for signals in data and in astronomy where generative methods can create galaxy models for dark energy experiments.
A Denoising Autoencoder.
Detecting signals in noisy channels is a very old problem. In the following paragraphs we consider the problem of identifying cosmic ray signals in radio astronomy. This work is based on “Classification and Recovery of Radio Signals from Cosmic Ray Induced Air Showers with Deep Learning”, M. Erdmann, F. Schlüter and R. Šmída 1901.04079.pdf (arxiv.org) and Denoising-autoencoder/Denoising Autoencoder MNIST.ipynb at master · RAMIRO-GM/Denoising-autoencoder · GitHub. We will illustrate a simple autoencoder that pulls the signals from the noise with reasonable accuracy.
For fun, we use three different data sets. One is a simulation of cosmic-ray-induced air showers that are measured by radio antennas as described in the excellent book “Deep Learning for Physics Research” by Erdmann, Glombitza, Kasieczka and Klemradt. The data consists of two components: one is a trace of the radio signal and the second is a trace of the simulated cosmic ray signal. A sample is illustrated in Figure 1a below with the cosmic ray signal in red and the background noise in blue. We will also use a second data set that consists of the same cosmic ray signals and uniformly distributed background noise as shown in Figure 1b. We will also look at a third data set consisting of the cosmic ray signals and a simple sine wave noise in the background Figure 1c. As you might expect, and we will illustrate, this third data set is very easy to clean.

Figure 1a, original data Figure 1b, uniform data Figure 1c, cosine data
To eliminate the nose and recover the signal we will train an autoencoder based on a classic autoencoder design with an encoder network that takes as input the full signal (signal + noise) and a decoder network that produces the cleaned version of the signal (Figure 2). The projection space in which the encoder network sends in input is usually much smaller than the input domain, but that is not the case here.

Figure 2. Denoising Autoencoder architecture.
In the network here the input signals are samples in [0, 1]500. The projection of each input consists of 16 vectors of length 31 that are derived from a sequence of convolutions and 2×2 pooling steps. In other words, we have gone from a space of dimension 500 to 496. Which is not a very big compression. The details of the network are shown in Figure 3.
The one dimensional convolutions act like frequency filters which seem to leave the high frequency cosmic ray signal more visible. To see the result, we have in figure 4 the output of the network for three sample test examples for the original data. As can be seen the first is a reasonable reconstruction of the signal, the second is in the right location but the reconstruction is weak, and the third is completely wrong.

Figure 3. The network details of the denoising autoencoder.

Figure 4. Three examples from the test set for the denoiser using the original data
The two other synthetic data sets (uniformly distributed random nose and noise create from a cosine function) are much more “accurate”.

Figure 5. Samples from the random noise case (top) and the cosine noise case (bottom)
We can use the following naive method to assess accuracy. We simply track the location of the reconstructed signal. If the maximum value of the recovered signal is within 2% of the maximum value of the true signal, we call that a “true reconstruction”. Based on this very informal metric we see that the accuracy for the test set for the original data is 69%. It is 95% for the uniform data case and 98% for the easy cosine data case.
Another way to see this result is to compare the response in the frequency domain, by applying an FFT to the input and output signals we see the results in Figure 6.

Figure 6a, original data Figure 6b, uniform data Figure 6c, cosine data
The blue line is the frequency spectrum of the true signal, and the green line is the recovered signal. As can be seen, the general profiles are all reasonable with the cosine data being extremely accurate.
Generative Autoencoders.
Experimental data drives scientific discovery. Data is used to test theoretical models. It is also used to initialize large-scale scientific simulations. However, we may not always have enough data at hand to do the job. Autoencoders are increasingly being used in science to generate data that fits the probabilistic density profile of known samples or real data. For example, in a recent paper, Henkes and Wessels use generative adversarial networks (discussed below) to generate 3-D microstructures for studies of continuum micromechanics. Another application is to use generative neural networks to generate test cases to help tune new instruments that are designed to capture rare events.
In the experiments for the remainder of this post we use a very small collection of images of Galaxies so that all of this work can be done on a laptop. Figure 7 below shows a sample. The images are 128 by 128 pixels with three color channels.

Figure7. Samples from the Galaxy image collection
Variational Autoencoders
A variational autoencoder is a neural network that uses an encoder network to reduce data samples to a lower dimensional subspace. A decoder network takes samples from this hypothetical latent space and uses it to regenerate the samples. The decoder becomes a means to map a uniform distribution on this latent space into the probability distribution of our samples. In our case, the encoder has a linear map from the 49152 element input down to a vector of length 500. This is then mapped by a 500 by 50 linear transforms down to two vectors of length 50. The decoder does a renormalization step to produce a single vector of length 50 representing our latent space vector. This is expanded by two linear transformations are a relu map back to length 49152. After a sigmoid transformation we have the decoded image. Figure 8 illustrates the basic architecture.

Figure 8. The variational autoencoder.
Without going into the mathematics of how this works (for details see our previous post or the “Deep Learning for Physics Research” book or many on-line sources), the network is designed so that the encoder network generates mean (mu) and standard deviation (logvar) of the projection of the training samples in the small latent space such that the decoder network will recreate the input. The training works by computing the loss as a combination of two terms, the mean squared error of the difference between the regenerated image and the input image and Kullback-Leibler divergence between the uniform distribution and the distribution generated by the encoder.
The Pytorch code is available as a the notebook in github. The same github directory contains the zipped datafile). Figure 9 illustrates the results of the encode/decode on 8 samples from the data set.

Figure 9. Samples of galaxy images from the training set and their reconstructions from the VAR.
An interesting experiment is to see how the robust the decoder is to changes in the selection of the latent variable input. Figure 10 illustrates the response of the decoder when we follow a path in the latent space from one instance from the training set to another very similar image.

Figure 10. image 0 and image 7 are samples from the training set. Images 1 through 6 are generated from points along the path between the latent variable for 0 and for 7.
Another interesting application was recently published. In the paper “Detection of Anomalous Grapevine Berries Using Variational Autoencoders” Miranda et.al. show how a VAR can be used to examine arial photos of vineyards to spot areas of possible diseased grapes.
Generative Adversarial Networks (GANs)
Generative Adversarial networks were introduced by Goodfellow et, al (arXiv:1406.2661) as a way to build neural networks that can generate very good examples that match the properties of a collection of objects.
As mentioned above, artificial examples generated by autoencoder can be used as starting points for solving complex simulations. In the case of astronomy, cosmological simulation is used to test our models of the universe. In “Creating Virtual Universes Using Generative Adversarial Networks” (arXiv:1706.02390v2 [astro-ph.IM] 17 Aug 2018) Mustafa Mustafa, et. al. demonstrates how a slightly-modified standard GAN can be used generate synthetic images of weak lensing convergence maps derived from N-body cosmological simulations. In the remainder of this tutorial, we look at GANs.
Given a collection r of objects in Rm, a simple way to think about a generative model is as a mathematical device that transforms samples from a multivariant normal distribution Nk(0,1) into Rm so that they look like they come from the actual distribution Pr. for our collection r. Think of it as a function

Which maps the normal distribution into a distribution Pg over Rm.
In addition, assume we have a discriminator function

With the property that D(x) is the probability that x is in our collection r. Our goal is to train G so that Pg matches Pr. Our discriminator is trained to reject images generated by the generator while recognizing all the elements of Pr. The generator is trained to fool the discriminator, so we have a game defined by minimax objective:

We have put a simple basic GAN from our previous post. Running it for many epochs can occasionally get some reasonable results as shown if Figure 11. While this looks good, it is not. Notice that it generated examples of only 3 of our samples. This repeating of an image for different latent vectors is an example of a phenomenon called modal collapse.

Figure 11. The lack of variety in the images is called model collapse.
acGAN
There are several variations on GANs that avoid many of these problems. One is called an acGAN for auxiliary classifier Gan developed by Augustus Odena, Christopher Olah and Jonathon Shlens. For an acGan we assume that we have a class label for each image. In our case the data has three categories: barred spiral (class 0), elliptical (class 1) and spiral (class 2). The discriminator is modified so that it not only returns the probability that the image is real, it also returns a guess at the class. The generator takes an extra parameter to encourage it to generate an image of the class. Let d-1 = the number of classes then we have the functions

The discriminator is now trained to minimize the error in recognition but also the error in class recognition. The best way to understand the details is to look at the code. For this and the following examples we have notebooks that are slight modifications to the excellent work from the public Github site of Chen Kai Xu from Chen Kai Xu Tsukuba University. This notebook is here. Figure 12 below shows the result of asking the generator to create galaxies of a given class. The G(z,0) generates good barred spirals, G(z,1) are excellent elliptical galaxies and G(z,2) are spirals.

Figure 12. Results from asgan-new. Generator G with random noise vector Z and class parameter for barred spiral = 0, elliptical = 1 and spiral = 2.
Wasserstein GAN with Gradient Penalty
The problem with modal collapse and convergence failure was, as stated above, commonly observed. The Wasserstein GAN introduced by Martin Arjovsky , Soumith Chintala , and L´eon Bottou, directly addressed this problem. Later Ishaan Gulrajani, Faruk Ahmed, Martin Arjovsky, Vincent Dumoulin and Aaron Courville introduced a modification called a Gradient penalty which further improved the stability of the GAN convergence. To accomplish this they added an additional loss term to the total loss for training the discriminator as follows:

Setting the parameter lambda to 10.0 this was seen as an effective value for reducing to occurrence of mode collapse. The gradient penalty value is computed using samples from straight lines connecting samples from Pr and Pg. Figure 13 shows the result of a 32 random data vectors for the generator and the variety of responses reflect the full spectrum of the dataset.

Figure 13. Results from Wasserstein with gradient penalty.
The notebook for wgan-gp is here.
Wasserstein Divergence for GANs
Jiqing Wu , Zhiwu Huang, Janine Thoma , Dinesh Acharya , and Luc Van Gool introduced a variation on WGAN-gp called WGAN-div that addresses several technical constraints of WGAN-gp having to do with Lipschitz continuity not discussed here (see the paper). They propose a better loss function for training the discriminator:

By experimental analysis the determine the best choice for the k and p hyperparameters are 2 and 6.
Figure 14 below illustrates the results after 5000 epochs of training. The notebook is here.

Figure 14. Results from WG-div experiments.
Once again, this seems to have eliminated the modal collapse problem.
Conclusion
This blog was written to do a better job illustrating autoencoder neural networks than our original articles. We illustrated a denoising autoencoder, a variational autoencoder and three generative adversarial networks. Of course, this is not the end of the innovation that has taken place in this area. A good example of recent work is the progress made on Masked Autoencoders. Kaiming He et. al. published Masked Autoencoders Are Scalable Vision Learners in December 2021. The idea is very simple and closely related to the underlying concepts used in Transformers for natural language processing like BERT or even GPT3. Masking simply removes patches of the data and trains the network to “fill in the blanks”. The same idea has been now applied to audio signal reconstruction. These new techniques show promise to generate more semantic richness to the results than previous methods.







 Fig 5. Microsoft FPGA based Project Brainwave [4, 5]
Fig 5. Microsoft FPGA based Project Brainwave [4, 5]









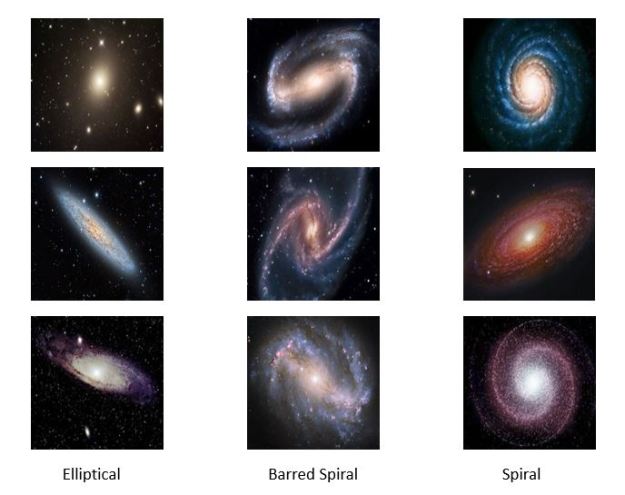

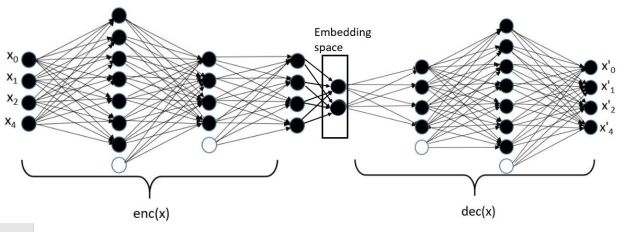


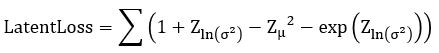











 Figure 7. Conceptual view of microservices as stateless services communicating with each other and saving needed state in distribute databases or tables.
Figure 7. Conceptual view of microservices as stateless services communicating with each other and saving needed state in distribute databases or tables.














 Figure 1. DCOS web interface.
Figure 1. DCOS web interface.















